Posts
(Re)Introducing Members by Interest
…Or how I learned to stop worrying and love having a database.
Last year I put together an application called Members by Interest to illustrate the particular interests of individual members of Congress by looking at the sorts of bills they sponsored and cosponsored.
At launch, the data driving the application was a series of (kinda large) static data files. This was great from a performance standpoint, but the process of creating those files involved having a Python script parse each Bill Status file from a given Congress before the files corresponding to that Congress could be written. When looking at the current Congress, this meant re-parsing hundreds (or thousands) of files every day that it was in session – a process that was not only inelegant and resource-intensive, but one that made automating updates difficult.
After some soul-searching (and confirming that the required database would fit within the GCP Free Tier constraints, I wrote an application to download Status files and read ‘em into a database. Once there, they can be queried, tallied up and fed to an API for consumption.
Of course re-writing the back-end meant some necessary changes to the front-end as well. This revision includes:
-
A cleaned-up UI and Display. Ranked lists now show a clearer distinction between high and low scoring members and there is more emphasis on party ID.
-
A revised method for computing the adjusted score that’s used for both ranking members and showing individual member charts. It now takes into account the distinction between being a cosponsor and an original cosponsor – the latter folks are the ones who are listed as cosponsors at introduction instead of having joined later, so there is a greater likelihood that they had input in constructing a given measure.
Having a database as the back-end instead of static files also means that the range of questions that can be posed to the data is expanded as well! Watch this space…
– ❉ –
Things may be slower than usual...
As noted in an earlier post , NCBI wants one to restrict calls to E-Utilities to no more than three per second.
If (like me) you are a developer who uses this interface on a regular basis, you may have noticed that NCBI has gotten a lot more aggressive about enforcing this limit. To wit, you will now start regularly getting an error if you exceed it from a given IP address. One way around this is to request an API key to sign requests with. This will get you 10 requests per second instead.
However this API key then applies to all applications (from all IP addresses) using it. It would still be possible for two or three simultaneous users with the same API key to exceed this limit and so lock access for everybody else. I am therefore modifying Search Workbench and the other pieces of Visualizing PubMed to only. call. every. 400. milliseconds (plus or minus a bit). You many notice a lag with some applications that make a lot of calls (such as Mapping MEDLINE ).
Do let me know if you encounter any problems with these applications…
– ❉ –
Are Narrow NOT Broad citations any good?
One critique I received on my presentation about what gets missed when only using the “Broad” version of the PubMed clinical queries was that maybe the citations missed were not “true positives”. That is, they might have characteristics in common that make them both invisible to the Broad query and also somehow less fit for purpose than their Narrow AND Broad fellows.
One relatively straightforward way to test this hypothesis is by using the work of the HIRU group at McMaster who originally developed the clinical queries. They curate a database called McMaster Plus. Articles in this database are carefully evaluated as to whether they meet the relevant inclusion criteria, so it can be presumed that they are “good”. Indeed, inclusion in this database was used as the test criteria for their 2012 revalidation study of the clinical queries. Therefore, one way to see whether or not Narrow NOT Broad citations are good ones is to see whether they show up in McMaster Plus.
Using some common clinical terms (such as “heart disease”, “breast cancer”, etc.) and the “Prognosis” category, I selected and downloaded a set of 1,021 Original Study citations from the McMaster Plus database via the Accessss interface. PMIDs were extracted from each citation, and these were searched against PubMed using the NCBI eutils, testing whether each one showed up under Prognosis/Broad, Prognosis/Narrow or Prognosis/Narrow NOT Prognosis/Broad.
The results were clear – many of these presumptively good citations exhibited the same behavior as noted in my original report. That is, they can be found using the Prognosis/Narrow clinical query but not when searching with Prognosis/Broad. Indeed, a little over 15% of the citations tested were missed when using Prognosis/Broad alone (Narrow OR Broad: 875, Narrow NOT Broad: 139). Whatever ways these Narrow NOT Broad citations may differ from those found using Broad alone, they cannot be assumed to be substandard.
– ❉ –
I was honorably mentioned!
I am pleased to announce that Members by Interest took “Honorable Mention” in the Congressional Data Challenge! Many thanks to the judges and the folks at the Library of Congress. Many congratulations to the winners as well!
To celebrate the recognition, I’ve added the ability to link directly to an individual search. For example, to see a ranked list of House members for Bills and Resolutions for the policy area ‘Health’: http://esperr.github.io/members-by-interest/?congress=115&chamber=house&subjecttype=policy&subject=Health
Or to see the particular interests of my own congressional delegation: https://esperr.github.io/members-by-interest/?congress=115&chamber=house&subjecttype=policy&member=H001071
More enhancements coming soon…
– ❉ –
Mind the Gap!
Another talk I presented at the 2018 Meeting of the Medical Library Association was on something I discovered while testing the hedges feature in Search Workbench. To wit, some searches using the “Narrow” version of a Clinical Query return results that are not seen when using the “Broad” version. This is particularly apparent when you examine the “Prognosis” queries:
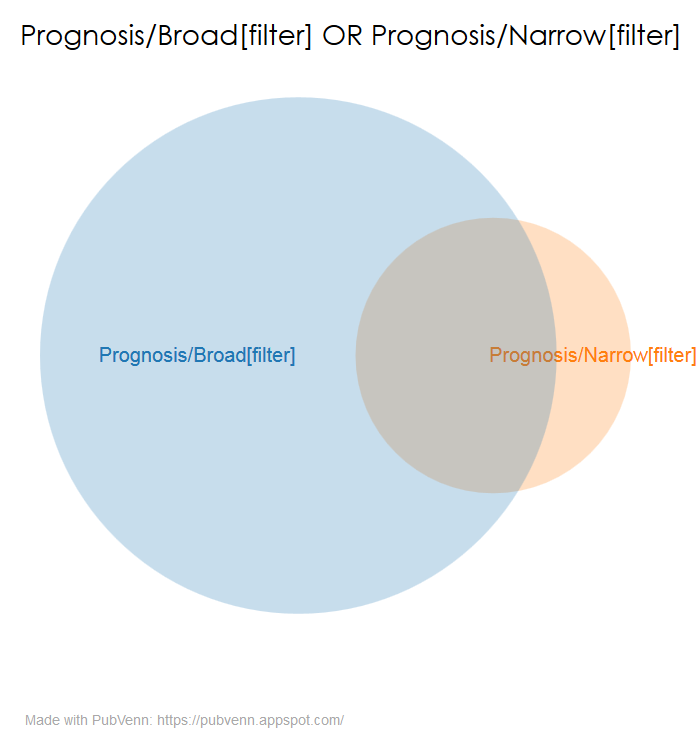
This seems problematic if the idea behind the “Broad” version is to show as much relevant material as possible. I’m in the process of writing this up in more detail, but in the meantime, please take a look at my slide deck and supporting materials (archived on osf):
Or if you have access to the MLA meeting materials, check out the video here.
– ❉ –